I replicated a paper on C. elegans chemotaxis but I'm not 100% happy with my work
So I was reading this paper and trying to replicate it because I guess that's what I do now. It is on a very simplified model of chemotaxis in C. elegans. I might get into details later, perhaps even a whole dedicated post if I get it working. But right now I just ran into something weird enough it deserves to be debugged and preserved here. Not because anyone will read it or find it helpful, just so I know what I've done. Now, there are two ways of simulating the movement. The more realistic way is by directly simulating neurons and having the change in the direction the worm is facing change based on the difference in voltage between two neurons. The other is to explicitly solve these differential equations in time and then extract what that same change in angle would be based on chemical gradient and what not. It seems very sensible to do either, provided you can actually analytically solve the equations which apparently the authors did. However, I am getting wildly conflicting results when I use one or the other. I'm 99% sure it is my fault but the authors never actually went and concerted the parameters in their neuron model to the derived parameters in the solved version. They did optimize their analytical solution to replicate chemotaxis but as far as I can tell they didn't simulate any neurons to do this. Well I tried to do that and when I then converted the biophysical parameters into the derived parameters I got nonsense.
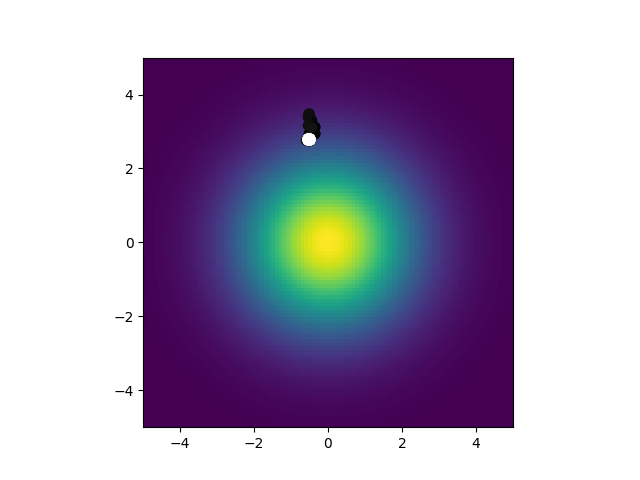
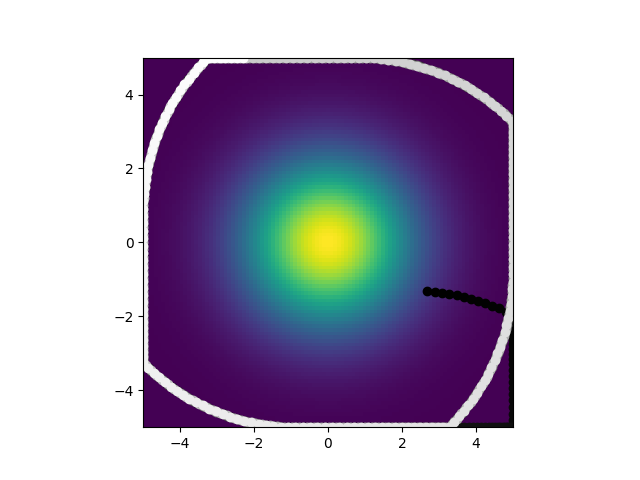
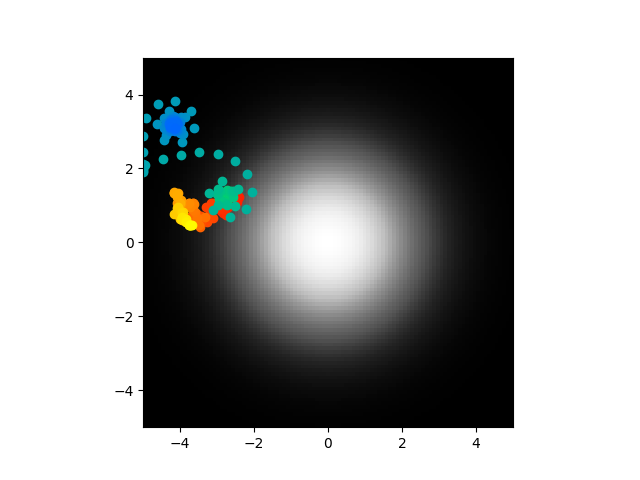
So what are you looking at? There is a gradient in the center that the worm is meant to seek. Points are also plotted for the worm's position. The darker a point the earlier it is in the simulation, the lighter, the later. So a white point is where the worm ended up, in this case after 15 simulated minutes. This was simulated with the neuron model given in the paper and random parameters for it. As you can see, not really great at center seeking. Not a huge surprise. What is surprising is this:
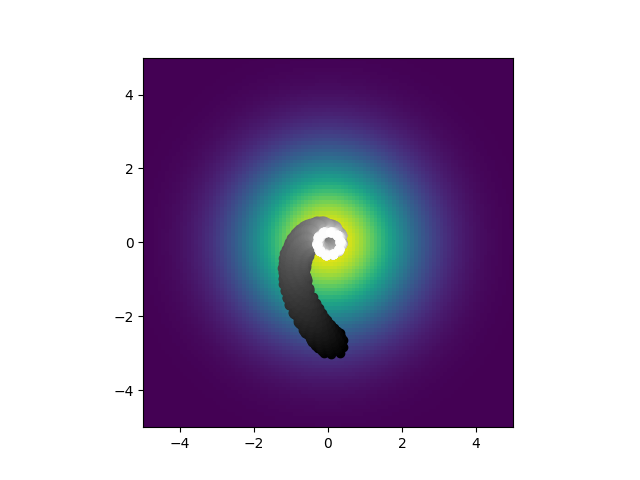
Using those same random initial conditions but converting them into the derived parameters necessary for the analytic solution, it looks nothing like what happens when you directly simulate it. It center seeks and forms these tight spirals. What is even weirder is that when I trained a model to optiomize the biophysical parameters, the ones for direct simualtion I got this:
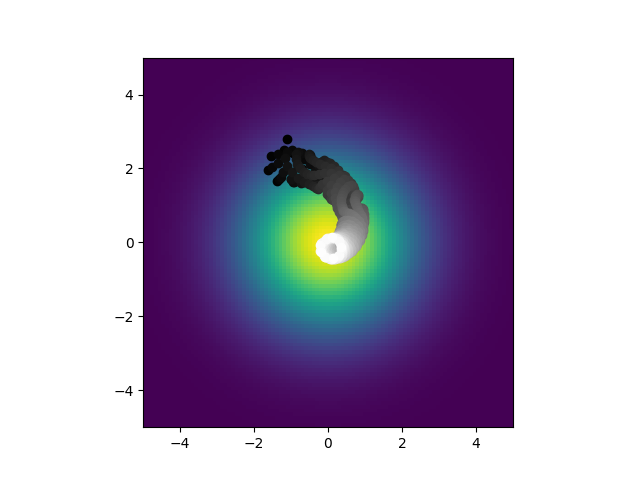
But this has a completely different set of biphysical parameters and is formed by direct simulation of neurons. But, when I convert it the derived parameters:
So I think my converter or my neuron simulation must be broken as they don't agree. Of course it is possible that the original paper made one too many simplifying assumptions but hey that's less likely than me just being wrong.
I'm going to explain more of what I did so far and publish everything I currently have because I really should take better notes.
The tl;dr is that this paper has
- non spiking neurons
- densely connected weight matrix
- biases on each neuron
- one sensory neurons that has an input proportional to the chemical concentration at that point
- only five neurons, the sensor, two for turning, and two hidden
- movement at a constant speed
Note, first mistake found, it is meant to have 6 neurons, there should be 3 hidden. I doubt this is the cause of everything falling to pieces but it speaks to my poor reading comprehension
The idea is that the neurons will be able to implicitly calculate the change in chemical gradient over time and change direction to move up that gradient. I first simulated this explicitly. In the paper it doesn't seem they ever do that. They further abstract it into a model that only takes into account the various time derivatives of the chemical gradient. This is equation 25 for those playing along at home. Different multiples of the different derivatives added up to be how fast the worm turns makes a very successful chemotaxis model. Yeah from our god's eye view we can just see where the concentration is strongest but if you can only sample one point and aren't physically large enough to even get the spatial gradient there it is kind of hard to tell how to move. Anyway, using the constants found in their paper you get graphs like this:
See how it doesn't perfect seek the center but still makes it there? See how it isn't this goofy spirally thing? I mean wow what a model, not many have made models like this let me tell you. Anyway my firs big idea is to have the voltage setpoint changed. I did -65mV because that's what human neurons are. But they have some equation (it isn't numbered but right before 16) to set the voltages automatically based on the synaptic weights, leak currents, and biases. I'll try that.
This, did not in fact, fix the issue. I am going to write some code to consolidate everything. I want it to show voltage over time, position of the both the neuron simulated worm and the time derivative thing one. That will really speed this bad boy up. Oh hey before I forget, I should talk about the end goal. So, I have seen some paper (not finding it right now) where a bias could be induced in C. elegans where they remembered where food was or something. Like you put them in a T shaped maze, put food in one arm, let them go for it. The after literally 1 trial, you could take them, put them in another T shaped maze but this time with no food and they would preferentially go to the side where the food was in the last maze. Simple memory/learning if you want to call it that. So that's why I want the biophysical model so much. I want to test learning rules on it and for that I need direct access to the neurons.
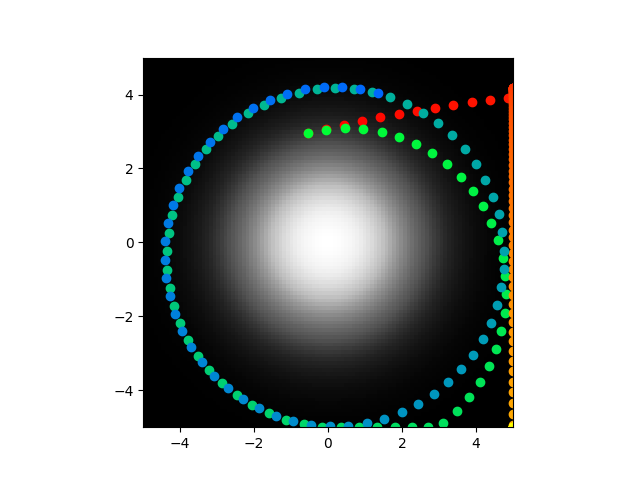
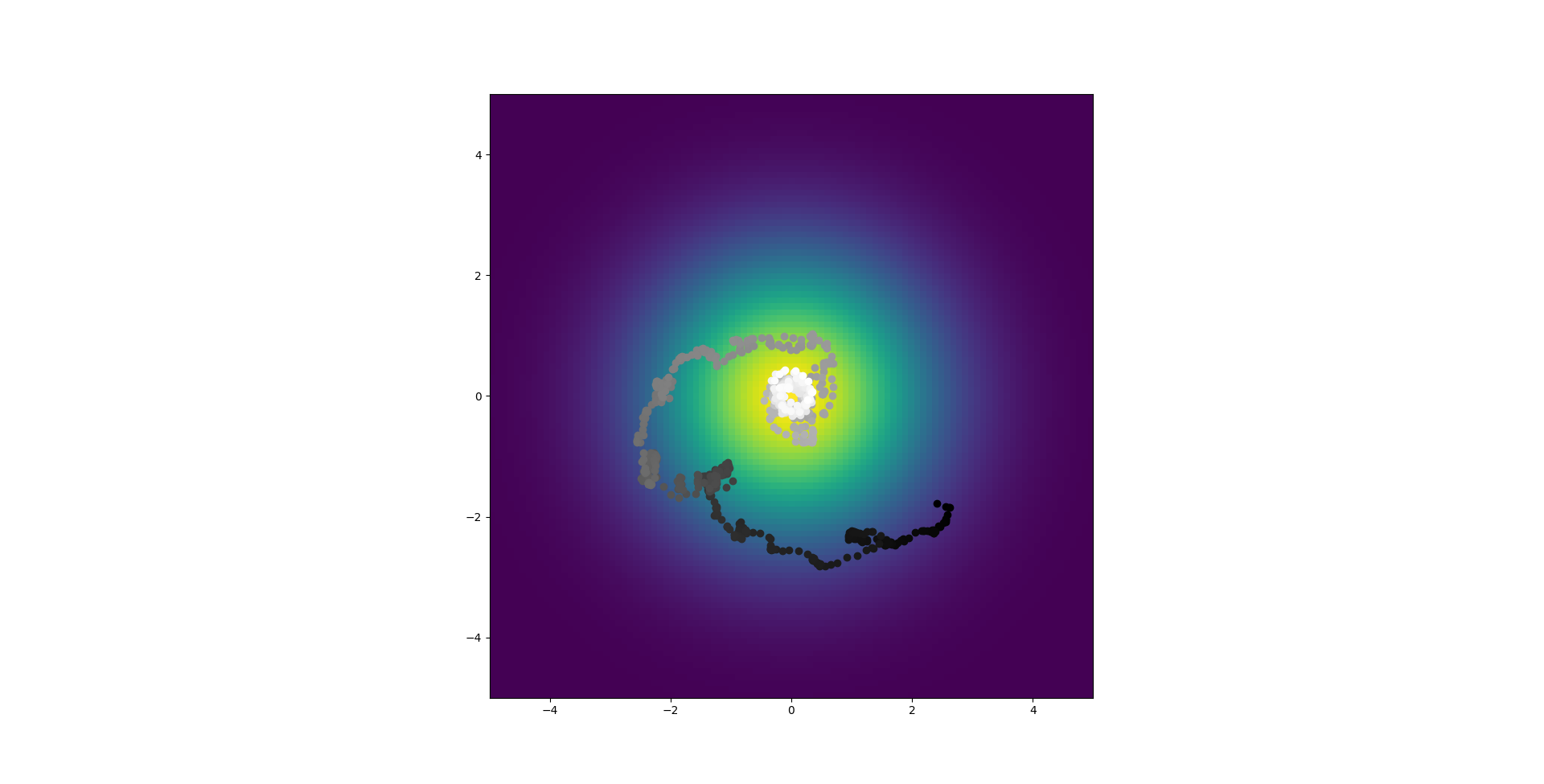
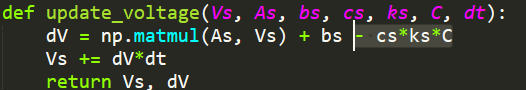
So the gradient is now grayscale, the abstract/analytical/time derivative based model (from here on out, the model based on equation 25 will be referred to as the abstract model and the model based on the voltages of explicitly simulated neurons will be the direct model) shows position with color that starts as red points and ends as yellow, the direct model goes from green to blue but it stayed essentially still. As you can see, disagreement still abounds. The code also shows the synaptic weights and the voltage over time but those aren't really relevant in this case. Code is pushed here. To be clear, these should perfectly overlap, or at leas be pretty close, that is not what is occurring.
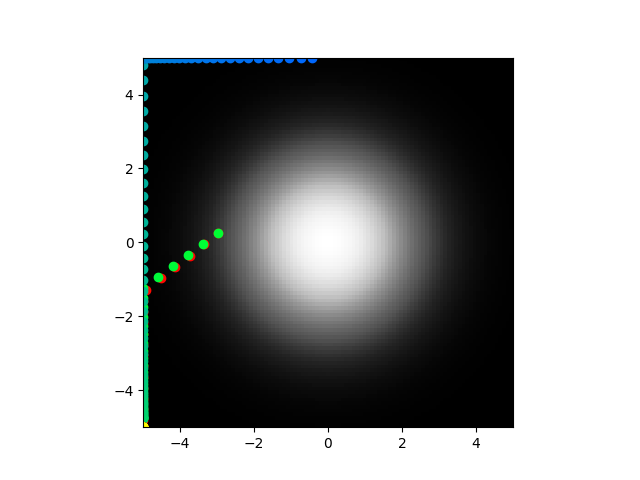
Ok so that is very encouraging. Actually maybe even a success. What did I do? Well I just swapped the sign of the chemical concentration contribution.
I am going to do some more testing on these same parameters.
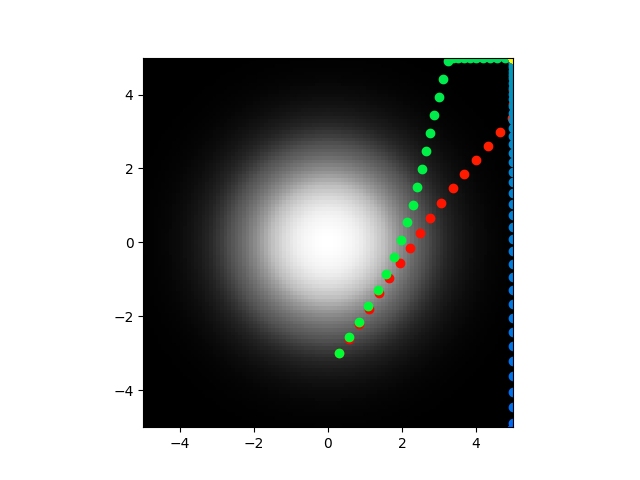
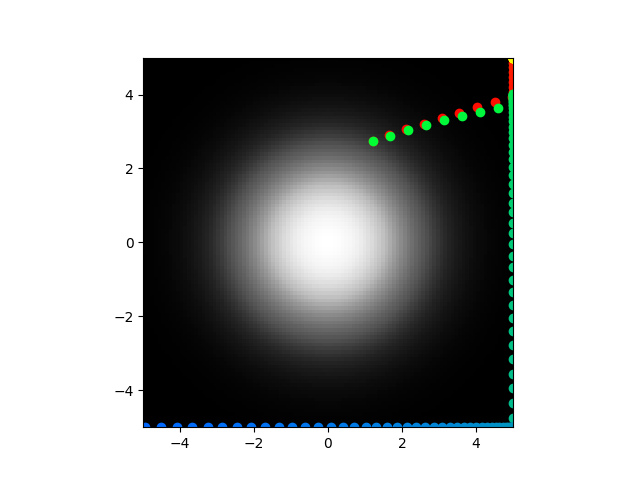
Yeah so clearly not perfect but the abstract model on has 3 terms so you can't expect it to follow exactly. This is great! Next step is to optimize the biophysical parameters so that I can recreate the abstract parameters and then I can see if the direct model will actually center seek. In my defense, if you take a look at equation 10 and 13 in the paper, there is no indication that that should be negative so I can't fault myself.
Just wanted to say that I'm not sure that negative sign matters, it seems to not always follow and performs about the same when it i positive, I think it was just luck.
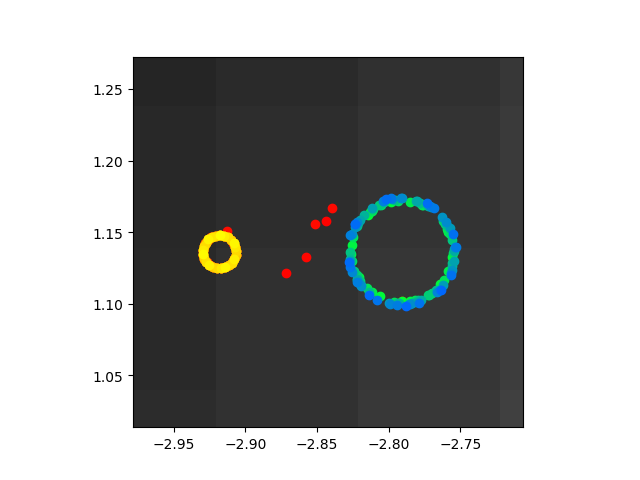
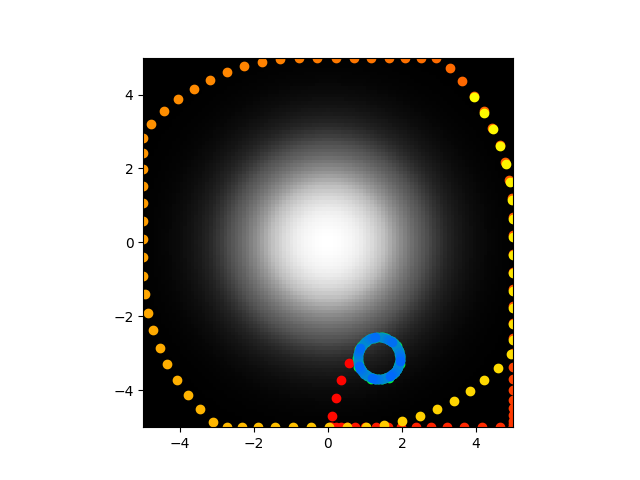
Ok so that above image is very zoomed in on the paths taken after I trained the biophysical parameters to minimize error between the resulting abstract parameters and those found in the paper. On the one hand, they paths are kind of close, at leas in the sense that they are both really highly localized. On the other hand, they are really bad because they don't center seek and are nowhere near the right values. Code here.
Tried this again with mean absolute error for the differences of the abstract parameters and also a term for the starting voltage, I want starting voltage low so that things don't blow up to infinity which can happen pretty easily. Garbage results, the abstract parameters are off by orders of magnitude and they paths aren't even close anymore.
Ugh I think things are either not saving or loading correctly. Yeah there's an off by one error somewhere in my code, I gotta track that down. Stupid bug messing up all my annealing work. Ok I'm 80% sure I fixed it, not 100% sure why it was happening because the change I made should have been inconsequential, I'm probably just bad at managing memory in Python.
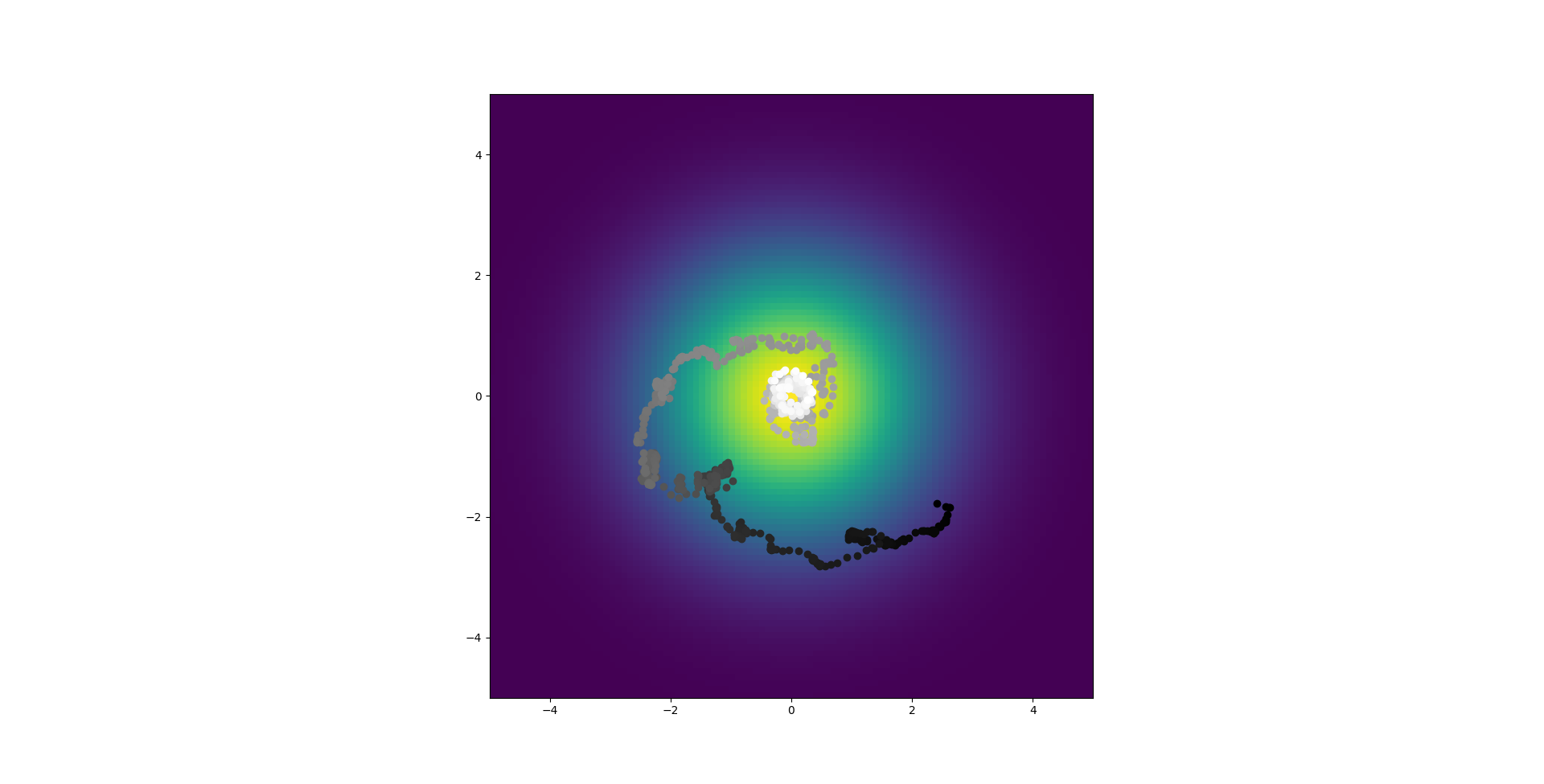
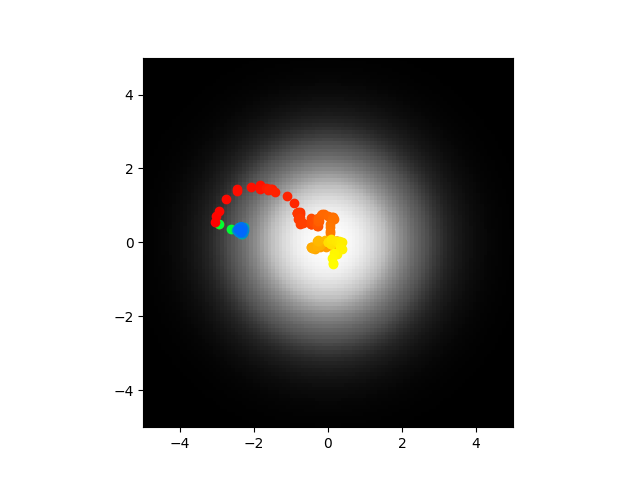
So two things, even with this issue fixed the simulated annealing still fails to reach parameters that replicate the chemotaxis also the direct and abstract model don't fit as closely as I thought which sucks. Yes those 3 initial examples were real but look at this:
Now maybe I could make them line up better but really I want to see if the chemotaxis works when the parameters are right, in that case I know they're wrong so it doesn't bother me. Of course if the trained model doesn't line up that would be a headache.
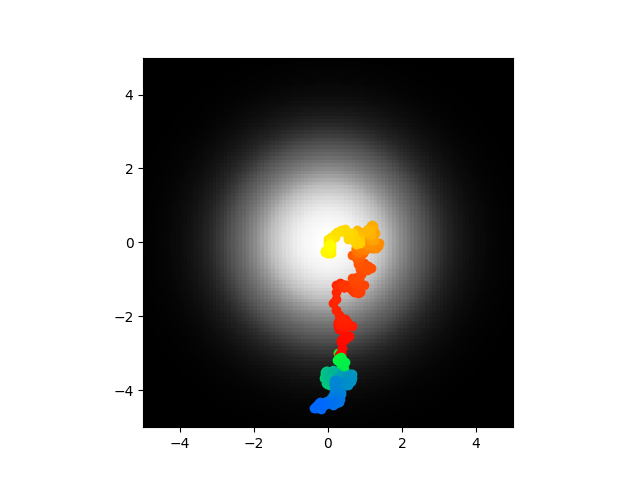
Hey so remember literally last sentence where I said that would be a headache? Well here we are, I got what looked like a pretty well optimized model I hade parameters set as 0.156, -2.674, 30.740, -95.298 (that's omega then z0, z1, z2) and optimal parameters should be 0, -0.562, 37.25, -88.27. It just didn't work, the direct model did not seek the center but the abstract model did indicating there is either a disconnect between theory and well less abstract theory or I did something stupid. No prizes for guessing which. Actually there is because I don't know yet. Here are the positions:
Code is here. Next steps? I really don't know. I guess confirm that the direct model and the abstract model line up, like I don't have bugs simulating my direct model. I know I had that alleged sign error, who knows how many more mistakes like that are in this code. Oh also I just tried retraining with the simulated annealing and it looks like that first run was some kind of fluke, I'm getting much higher error now and the voltages are all over the place, they run off to infinity which makes me sad.
Ugh I'm such an idiot. I wasn't updating the chemical concentration strength for the two things independently so the direct model had the chemical strength for the abstract model and it's all messed up. I fixed it now but I still have to retest stuff. Ok, I retested and it still doesn't work with the annealing but uh I'll keep working I guess. Code is here.
Honestly it seems more coincidental than anything whether or not the direct and the abstract simulation line up. I don't know what to do with that. The voltages blow up to infinity in these simulation. I'm going to add another term to the anneal to try and prevent that but it will mean explicitly simulating neurons anyway and I don't know if it will even fix the issue. That is a tomorrow issue dude I'm so tired. Oh I tried training without changing the k parameter which is sense strength, it didn't ever get good conditions. See I have a suspicion that the sense strength was too high because it was like an order of magnitude greater than the initial value but I can't just fix it apparently.
Tried that, unsurprisingly it did not work. I think effort should be focused on making sure the direct and abstract simulations agree in the untrained cases because if they don't in that case why should they in the trained case? We already have pretty decent chemotaxis with abstract parameters derived from the biophysical ones, the direct simulation just doesn't work with those.
Ok so with some pretty vibes based testing, the b parameter in the direct model being set to non zero values makes the models diverge quite a bit so I'm going to train with the constraint that b must be 0 for all neurons. This might be because b being 0 forces omega to also be 0 which is what we want but hey who knows.
This did a little thing I like to call 'not work'.
Chemotaxis on abstract is good but still garbage for the direct model. I'm thinking that k should be smaller again because it was again orders of magnitude too high.
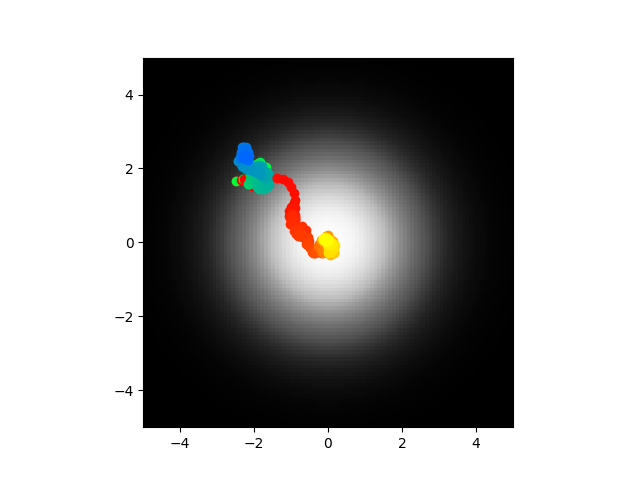
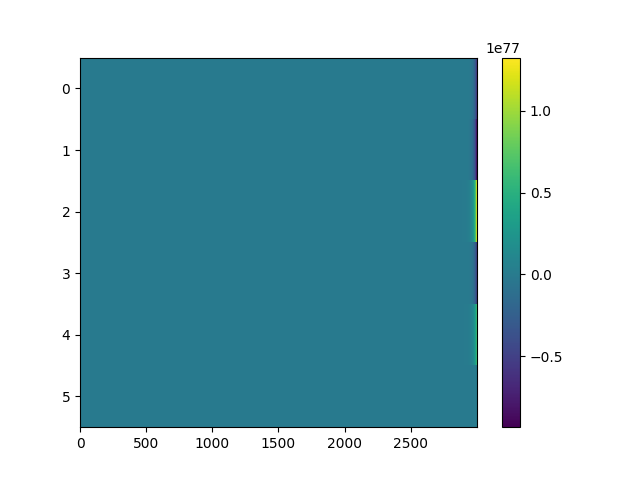
Another result where the abstract chemotaxis is fine and the direct model is garbage. The second image is the voltage. See how it's absolutely crazy? I don't know how to fix that.
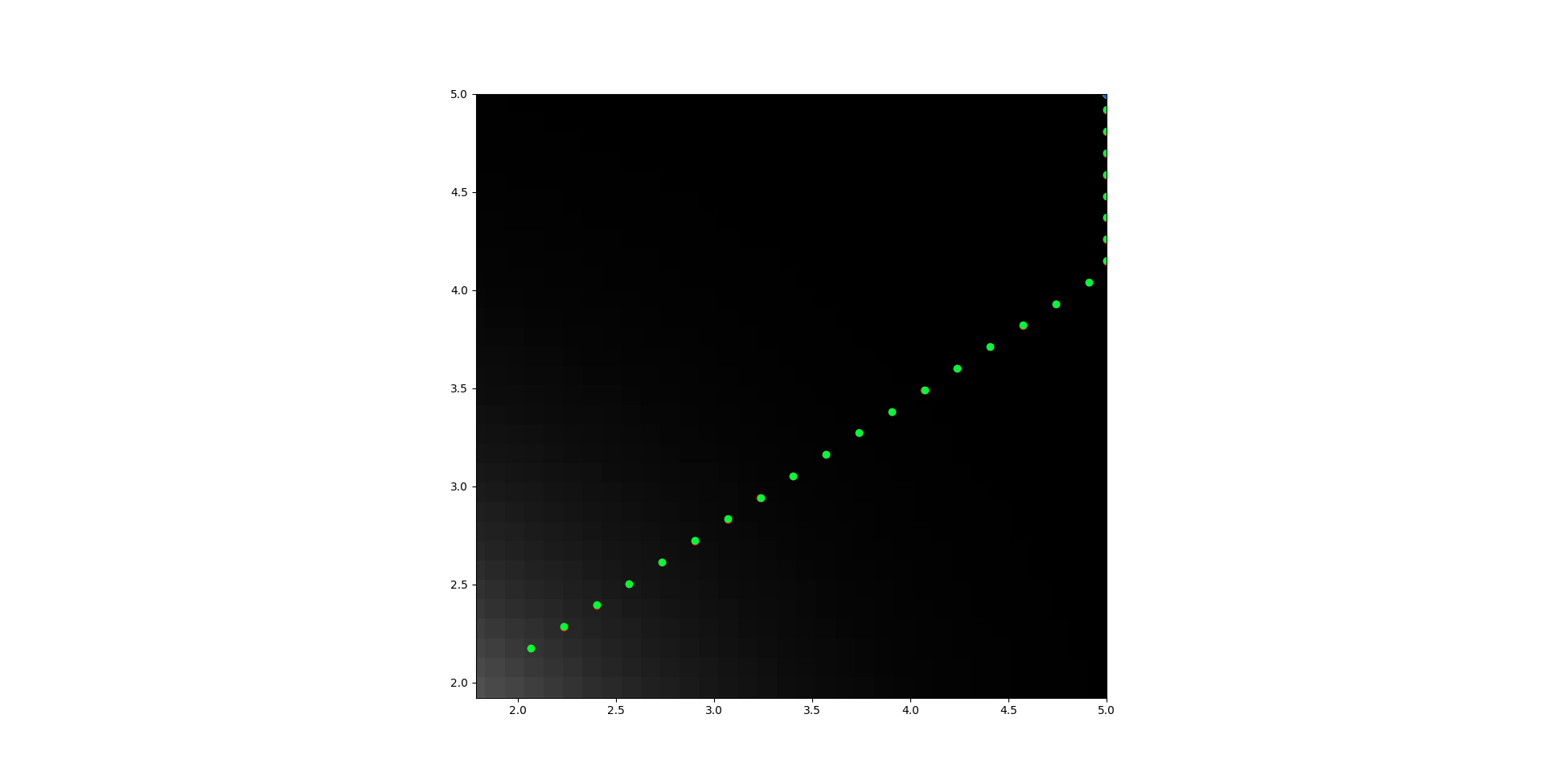
I realized that I never showed what happen when b = 0. That image is perhaps the best example. I'm going to show a couple others that are less good.
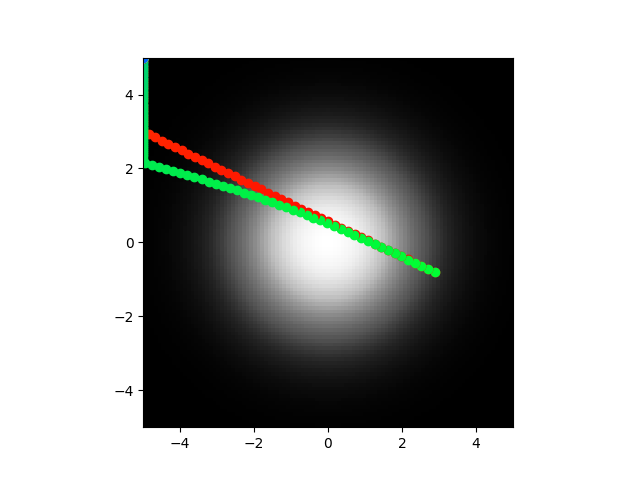
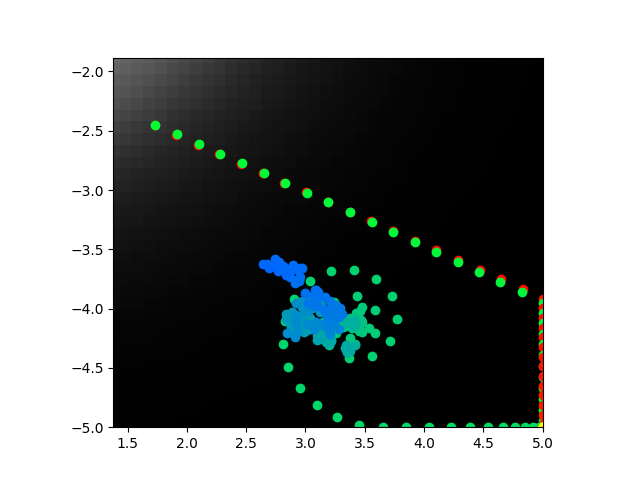
Ok so I trained with optimization for the normal parameters and also keeping the voltage low. It kind of worked? It is promising enough I'm going to retrain a lot and see what happens. Sometimes the direct model center seeks, sometimes the abstract model does. They don't line up super close and aren't really highly correlated when they do or don't. But the voltage is low always so maybe I can tune the error function and get good reliable behavior for both.
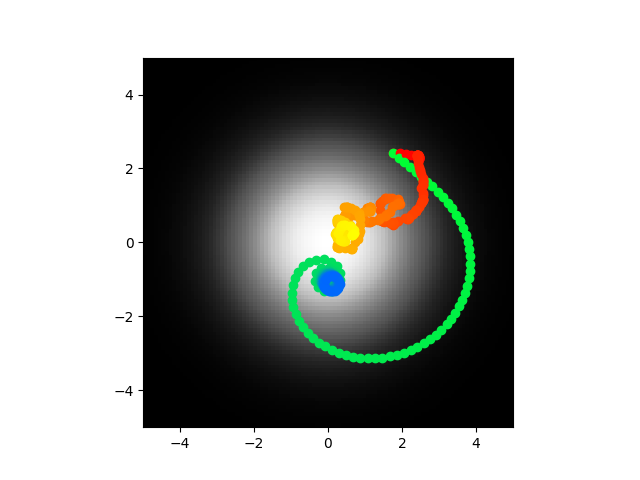
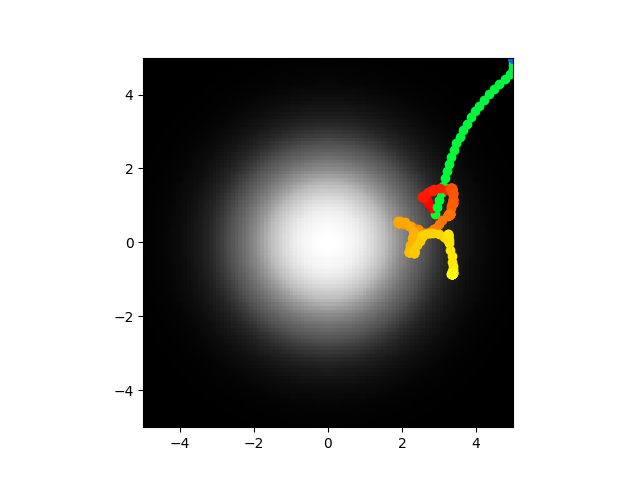
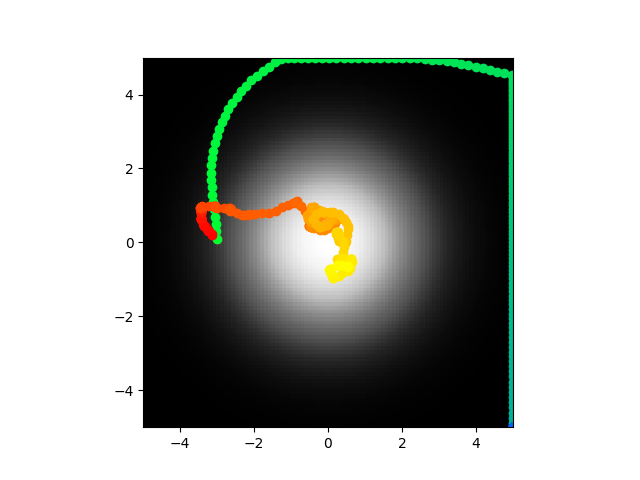
Code here. Going to train on the same thing again pretty much.
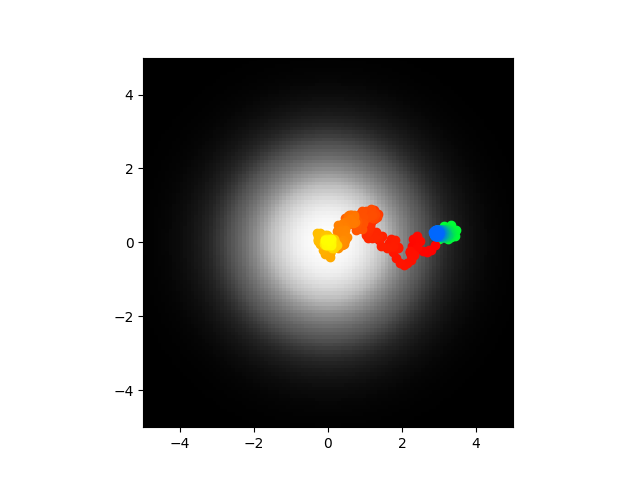
Modified the error function slightly to only punish if the absolute value of voltage was over 100mV. The abstract model works but clearly the direct doesn't. I'm thinking of limiting the k value again and retraining. Code here.
Also I might try normalizing the values some how. z0 and z2 are different orders of magnitude, perhaps that is why training is not working well.
So I tried that and uh it doesn't work. It does have a really high agreement between direct and abstract models. Maybe that is overselling it, they go in the same direction broadly speaking at the start. Example:
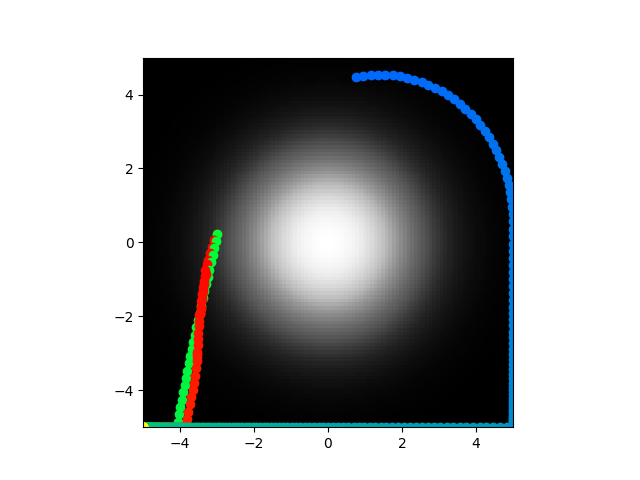
But yeah, still doesn't center seek so time to do my favorite thing: hyperparmeter tuning on a training algorithm! I swear all my projects lead back to this I'm so tired of it. Code here. I will now begin a bunch of tiny modifications.
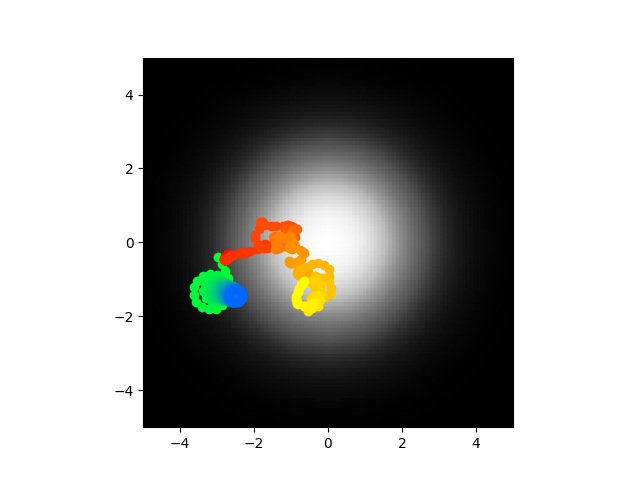
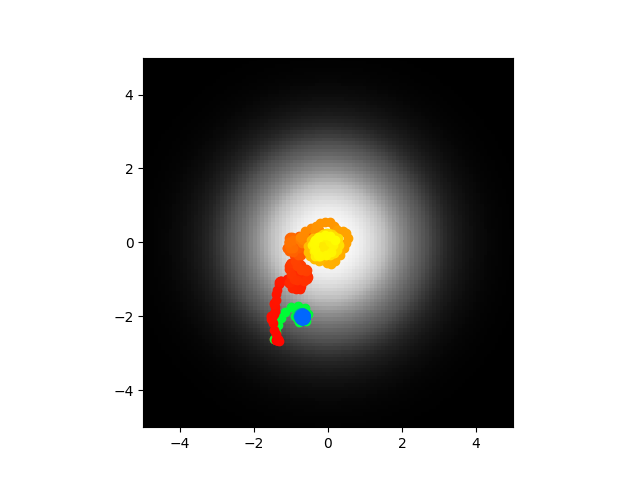
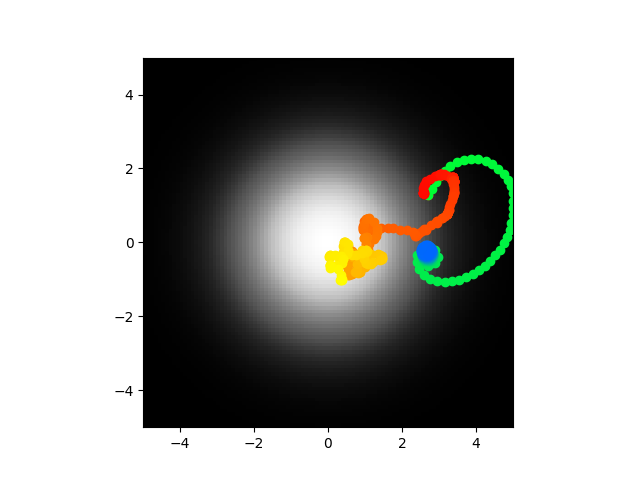
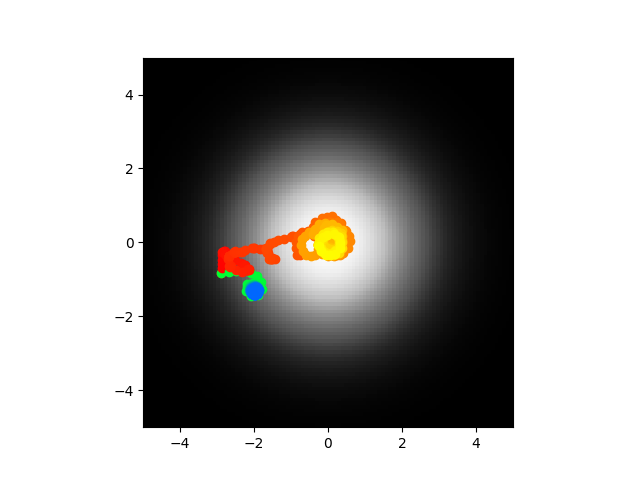
Ok cool so yeah I did this and I got to say I think I'm going to call it. The voltages are stable as in they don't blow up to infinity, the synaptic weights are reasonable values, the abstract model center seeks most of the time. It seems like the most logical explanations at this point are either 1) my code is bugs somewhere and the biophysical/direct model is not accurate or 2) the abstract model is too abstract and looses something important. I am going to run this again and if it doesn't work I think I'm going to call it or at least pause and work on something else. Code of course.
Yeah I think this is shot. Just for a second I thought hey maybe my velocity setting is wrong or maybe gamma but they used 0.022 for velocity, I had 0.02, as for gamma they optimized it with the simulated annealing which is what I'm doing. This is kind of demoralizing man. Did my half fail or is there really a disconnect? It is so tough to say. I should probably check all the indexing to make sure I didn't just like transpose a matrix on accident or something but honestly I'd be surprised if that was the reason. If there is no update, assume I didn't check or there were no problems there.